男女共同参画学協会連絡会
支援企業による広告記事
- ソーラボジャパン株式会社
- CAD を使って光学装置を設計してみよう
- 「生物物理」2025年10月号
- ソーラボジャパン株式会社
- 次世代2光子顕微鏡―小型化がもたらす新たな可能性
- 「生物物理」2024年10月号
- ソーラボジャパン株式会社
- サイエンティフィックカメラと周辺機器の同期
- 「生物物理」2023年10月号
- ソーラボジャパン株式会社
- 顕微鏡のリノベーション ~ 顕微鏡ポートを活用した光学系の導入
- 「生物物理」2022年12月号
「Biophysics and Physicobiology」に Shigeyuki Matsumoto, Shoichi Ishida, Kei Terayama, Yasuhshi Okuno による "Quantitative analysis of protein dynamics using a deep learning technique combined with experimental cryo-EM density data and MD simulations" をJ-STAGEの早期公開版として掲載
2023年05月16日 学会誌
日本生物物理学会欧文誌[Biophysics and Physicobiology]に以下の論文が早期公開されました。
Shigeyuki Matsumoto, Shoichi Ishida, Kei Terayama, Yasuhshi Okuno
"Quantitative analysis of protein dynamics using a deep learning technique combined with experimental cryo-EM density data and MD simulations"
URL:https://doi.org/10.2142/biophysico.bppb-v20.0022
- Abstract
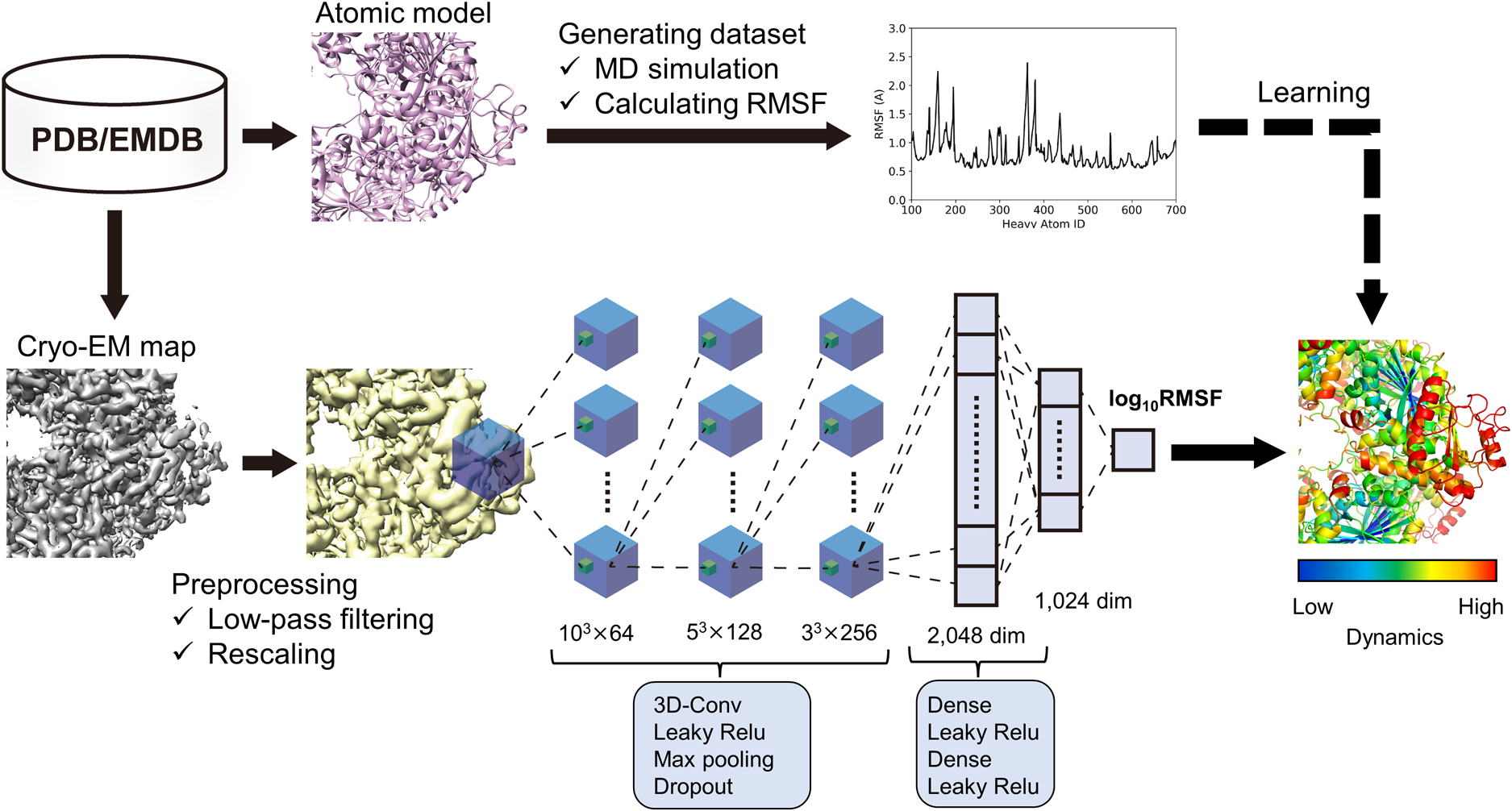
- Protein functions associated with biological activity are precisely regulated by both tertiary structure and dynamic behavior. Thus, elucidating the high-resolution structures and quantitative information on in-solution dynamics is essential for understanding the molecular mechanisms. The main experimental approaches for determining tertiary structures include nuclear magnetic resonance (NMR), X-ray crystallography, and cryogenic electron microscopy (cryo-EM). Among these procedures, recent remarkable advances in the hardware and analytical techniques of cryo-EM have increasingly determined novel atomic structures of macromolecules, especially those with large molecular weights and complex assemblies. In addition to these experimental approaches, deep learning techniques, such as AlphaFold 2, accurately predict structures from amino acid sequences, accelerating structural biology research. Meanwhile, the quantitative analyses of the protein dynamics are conducted using experimental approaches, such as NMR and hydrogen-deuterium mass spectrometry, and computational approaches, such as molecular dynamics (MD) simulations. Although these procedures can quantitatively explore dynamic behavior at high resolution, the fundamental difficulties, such as signal crowding and high computational cost, greatly hinder their application to large and complex biological macromolecules. In recent years, machine learning techniques, especially deep learning techniques, have been actively applied to structural data to identify features that are difficult for humans to recognize from big data. Here, we review our approach to accurately estimate dynamic properties associated with local fluctuations from three-dimensional cryo-EM density data using a deep learning technique combined with MD simulations.
URL: https://doi.org/10.2142/biophysico.bppb-v20.0022